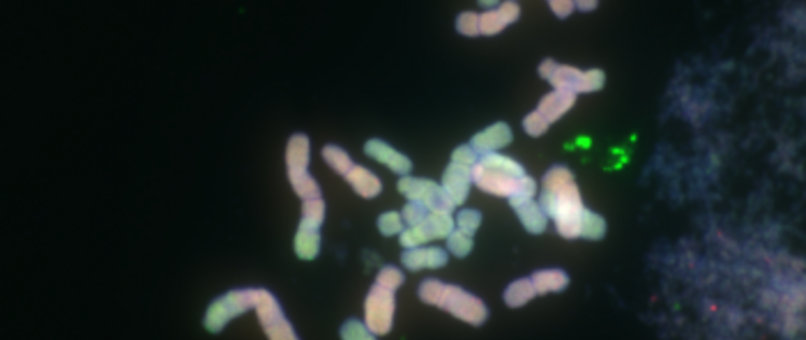
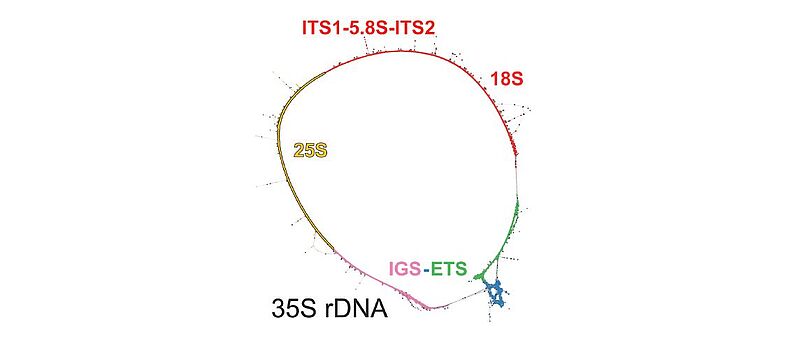
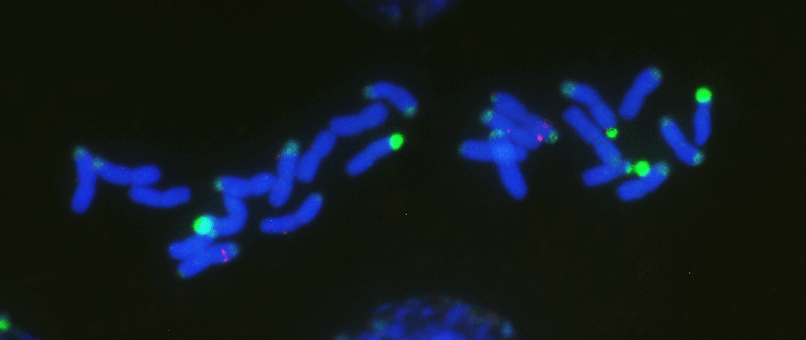
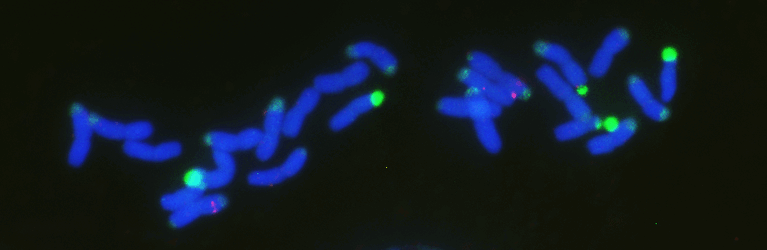
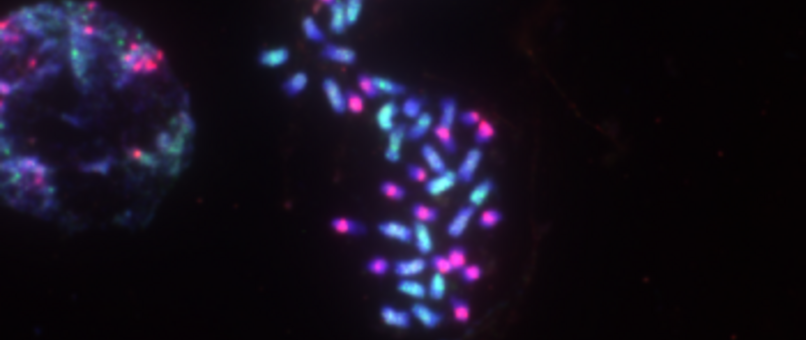
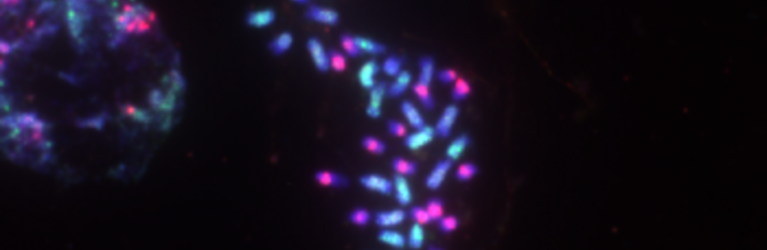
Our research focuses on plant genome evolution, ranging from the evolution of plant chromosome numbers and incidence of polyploidy, to detailed analyses of the composition of plant chromosomes and genomes. Of special interest are analyses of the dynamics of genomic changes following hybridization and polyploidization. Our research includes aspects of phylogeny and systematics, as well as genomics and genetics. We focus explicitly, but not exclusively, on the evolution of the repetitive DNA fraction of the genome which encompasses dispersed transposable elements and various tandem repeats such as telomeric motifs, rDNAs and satellite DNAs.
Chromosome number and structure dynamics are important to speciation and evolution. Application of high-throughput sequencing technologies in concert with fluorescence in situ hybridization allows us to study chromosome evolution in systems with highly dynamic chromosome numbers. Of special interest are supernumerary B chromosomes (Bs) and supernumerary chromosomal segments (SCSs).
Polyploidy is particularly important in plant evolution. We focus on identifying parental origin of polyploids and use a comparative approach to study genome evolution following auto- and allopolyploidization. We also develop novel approaches to infer the age of allopolyploids, which are used for calibrating the dynamics of the evolution of their repetitive DNA fraction.
Repetitive DNA represents the most dynamic fraction of plant genomes. Both dispersed transposable elements and tandemly repeated satellite DNAs are important drivers of genome size and chromosome structure evolution in plants. Their dynamic evolution is correlated with high levels of divergence even among among closely related taxa. We utilize high-throughput sequencing and graph-based clustering approaches, in addition to experimental approaches, for characterization and inferring the patterns of the evolution of repetitive elements in various plant genomes.
Our research is also reflected in teaching at bachelor, master and PhD levels.
Recent Publications
Groot Crego C, Hess J, Yardeni G, de la Harpe M, Priemer C, Beclin F, Saadain S, Cauz-Santos L, Temsch E, Weiss-Schneeweiss H, Barfuss M, Till W, Weckwerth W, Heyduk K, Paun O, Leroy T. (2024) CAM evolution is associated with gene family expansion in an explosive bromeliad radiation. The Plant Cell (in press)
Palombo NE, Weiss-Schneeweiss H, Carrizo Garcia C. (2024) Evolutionaty relationships, hybridization and diversification under domestication of the locoto chile (Capsicum pubescens) and its wild relatives. Frontiers in Plant Science 15: 1353991.
Taraska V, Duchoslav M, Hrones M, Batousek P, Lamla F, Temsch EM, Weiss-Schneeweiss H, Travnicek B. (2024) Dactylorhiza maculata agg. (Orchidaceae) in Central Europe: intricate patterns in morphological variability, cytotype diversity and ecology support the signle species concept. Folia Geobotanica 58: 151-188.
Weiss-Schneeweiss H, Jang T-S (2023) Formamide-free Genomic In Situ Hybridization (ffGISH). In: Plant Cytogenetics and Cytogenomics: Methods and Protocols; Heitkam T, Garcia S. (eds.). Methods in Molecular Biology, vol. 2672, Springer, pp. 257-264.
Kim H, Chi B, Lee C, Paik J-H, Jang C-G, Weiss-Schneeweiss H, Jang T-S (2023) Does the evolution of micromorphology accompany chromosomal changes on dysploid and polyploid levels in the Barnardia japonica complex (Hyacinthaceae)? BMC Plant Biology 23: 485.2022
Greimler J, Temsch EM, Xue Y, Weiss-Schneeweiss H Volkova P, Peintinger M, Wasowicz P, Shang H, Schanzer I, Chiapella JO (2022) Genome size variation in Deschampsia caespitosa sensu lato (Poaceae) in Eurasia. Plant Systematics and Evolution 308: 9.
Senderowicz M, Nowak T, Weiss-Schneeweiss H, Papp L, Kolano B (2022) Molecular and cytogenetic analysis of rDNA evolution in Crepis sensu lato. International Journal of Molecular Sciences 23: 3643.
Eriksson MC, Mandakova T, McCann J, Temsch EM, Chase MW, Hedren M, Weiss-Schneeweiss H, Paun O (2022) Repeat dynamics across timescales: a perspective from sybling allotetraploid marsh orchid (Dactylorhiza majalis s.l.). Molecular Biology and Evolution 39: msac167.
Park I, Choi B, Weiss-Schneeweiss H, So S, Myeong H-H, Jang T-S (2022) Comparative analyses of complete chloroplast genomes and karyotypes of allotetraploid Iris koreana and its putative diploid parental species (Iris series Chinenses, Iridaceae). International Journal of Molecular Sciences 23: 10929.
Carrizo Garcia C, Barbosa GE, Palombo N, Weiss-Schneeweiss H (2022) Diversification of chiles (Capsicum, Solanaceae) through time and space: new insight from genome-wide RAD-seq data. Frontiers in Genetics 13: 1030536.
Taraska V, Batousek P, Duchoslav M, Temsch EM, Weiss-Schneeweiss H (2021) Morphological variability, cytotype diversity, and cytogeography of populations traditionally calles Dactylorhiza fuchsii in Central Europe. Plant Systematics and Evolution 307: 51.
Muhamad A, Leroy T, Krigas N, Temsch EM, Weiss-Schneeweiss H, Lexer C, Sehr EM, Paun O (2021) Spatial and ecological drivers of genetic structure in Greek populations of Alkanna tinctoria (Boraginaceae), a polyploid medicinal herb. Frontiers in Plant Science 12: 706574.
Hartmann J, Silbernagl L, Schneeweiss GM, Barfuss MHJ, Weiss-Schneeweiss H, Schönswetter P (2021) Euphrasia ultima, a new locally endemic diploid species from the Ortler / Ortles range (Italy), is a close relative of widespread allotetraploid E. minima. Plant Biosystems 156: 893-907.
Lysak MA, Weiss-Schneeweiss H (2021) Editorial: Chromosomal Evolution in Plants. Frontiers in Plant Science 12: 726330.
Senderowicz M, Nowak T, Rojek-Jelonek M, Bisaga M, Papp L, Weiss-Schneeweiss H, Kolano B. (2021) Descending dysploidy and bidirectional changes in genome size accompanied Crepis (Asteraceae) evolution. Genes 12: 1436.
McCann J, Macas J, Novak P, Stuessy TF, Villaseñor J, Weiss-Schneeweiss H (2020) Differential genome size and repetitive DNA evolution in diploid species of Melampodium sect. Melampodium (Asteraceae). Frontiers in Plant Science 11: 362. DOI: 10.3389/fpls.2020.00362.
Choi B, Weiss-Schneeweiss H, Temsch E, So S, Myeong HH, Jang TS (2020) Genome size and chromosome number evolution in Korean Iris L. species. Plants 9: 1284. DOI: 10.3390/plants9101284.
Stuessy TF, Weiss-Schneeweiss H (2019) What drives polyploidization in plants? New Phytologist 223: 1690–1692. DOI: 10.1111/nph.15929.
Mlinarec J, Skuhala A, Jurković A, Malenica N, McCann J, Weiss-Schneeweiss H, Bohanec B, Besendorfer V. (2019) The repetitive DNA composition in the natural pesticide producer Tanacetum cinerariifolium: interindividual variation of subtelomeric tandem repeats. Frontiers in Plant Science 10:613.
Urtubey E, Tremetsberger K, Baeza CM, López-Sepúlveda P, König C, Samuel R, Weiss-Schneeweiss H, Stuessy TF, Ortiz MA, Talavera S, Terrab A, Ruas CF, Matzenbacher NI, Muellner AN, Guo Y-P (2019) Systematics of Hypochaeris section Phanoderis (Asteraceae, Cichorieae). Systematic Botany Monographs 106.
For more see PUBLICATIONS